Paul M. Lieberman, Ph.D.
-
Hilary Koprowski, M.D., Endowed Professor and Program Leader, Gene Expression and Regulation Program, Ellen and Ronald Caplan Cancer Center
-
Director, Center for Chemical Biology & Translational Medicine
Lieberman studies how certain viruses establish a long-term latent infection that can lead to cancer or autoimmune disorders.
Lieberman joined The Wistar Institute in 1995 as an assistant professor. He earned his bachelor’s degree in chemistry from Cornell University and a doctorate in pharmacology/virology from The Johns Hopkins University School of Medicine, which was followed by a postdoctoral fellowship at the University of California, Los Angeles.
Lieberman Chairs the Program in Gene Expression and Regulation at the Ellen and Ronald Caplan Cancer Center at The Wistar Institute. In 2010, Lieberman became the first director of The Wistar Institute Center for Chemical Biology and Translational Medicine. Using the advanced screening technologies of Wistar’s Molecular Screening Facility, the Center enables scientists to identify and characterize new molecules and compounds that hold the most promise for developing into therapeutic drugs for cancer and other diseases.
The Lieberman Laboratory

The Lieberman Laboratory
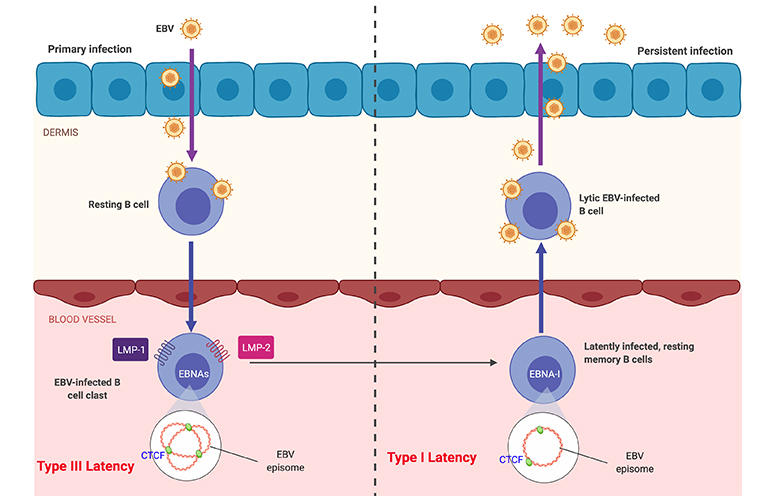
Research in the Lieberman laboratory centers on understanding how the cancer-associated viruses persist in a latent state and increase the risk of cancer and autoimmune disorders. EBV and KSHV establish latent infections that are associated with several human malignancies, including Burkitt’s lymphoma, nasopharyngeal carcinoma, Hodgkin’s disease, and post-transplant lymphoproliferative disorder for EBV, and Kaposi’s Sarcoma for KSHV. EBV has also been implicated in multiple sclerosis and other autoimmune disorders.
Lieberman and his team found that oncogenic viruses can interact with cellular proteins that regulate telomeres—the repetitive DNA sequences found at the ends of chromosomes. Telomeres protect chromosomes from loss of genetic information, and a similar process is thought to preserve the virus during latency. The Lieberman laboratory worked out several biochemical pathways that control the stability, replication, and gene expression patterns of the latent virus. New research also focuses on the epigenetic controls of latent viruses and human telomeres, and how interactions between viruses and telomeres may induce a malignant transformation of the infected cell.
-
Senior Staff Scientists
Troy Messick, Ph.D.
Samantha Soldan, Ph.D. -
Associate Staff Scientists
Amit Gurav, Ph.D.
Urvi Zankharia, Ph.D. -
Postdoctoral Fellows
Leonardo Josue Castro-Munoz, Ph.D.
Chris Chen, Ph.D.
Chenhe Su, Ph.D. -
Research Assistants
Asim Ashgar
Andreas Wiedmer
Olga Vladimirova
Research
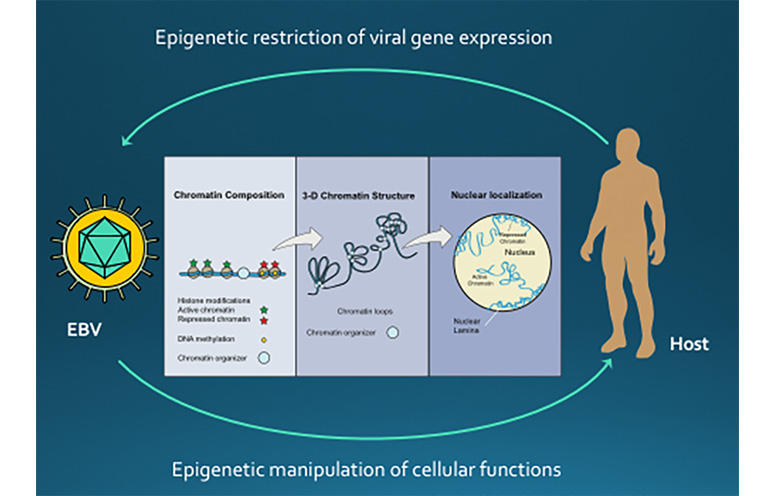
EPIGENETIC CONTROL OF VIRAL LATENCY
Research in the Lieberman laboratory centers on understanding how the cancer-associated viruses, like Epstein-Barr virus (EBV) and Kaposi’s Sarcoma Associated Herpesvirus (KSHV), persist in a latent state and increases the risk of cancer cell evolution. EBV and KSHV establish latent infections that are associated with several human malignancies, including Burkitt’s lymphoma, nasopharyngeal carcinoma, Hodgkin’s disease, and post-transplant lymphoproliferative disorder for EBV, and Kaposi’s Sarcoma for KSHV.
The researchers have recently found that viral DNA replication and maintenance is regulated by interactions with cellular telomere binding proteins. Telomeres are the repetitive DNA sequences found at the ends of chromosomes. Telomeres protect chromosomes from loss of genetic information, and a similar process is thought to preserve the virus during latency. The Lieberman research team has worked out several biochemical pathways that control the stability, replication, and gene expression patterns of the latent virus. They have found that changes in viral chromatin structure alters the cancer-risk associated with latent infection.
VIRUS MODULATION OF HOST CHROMOSOMES
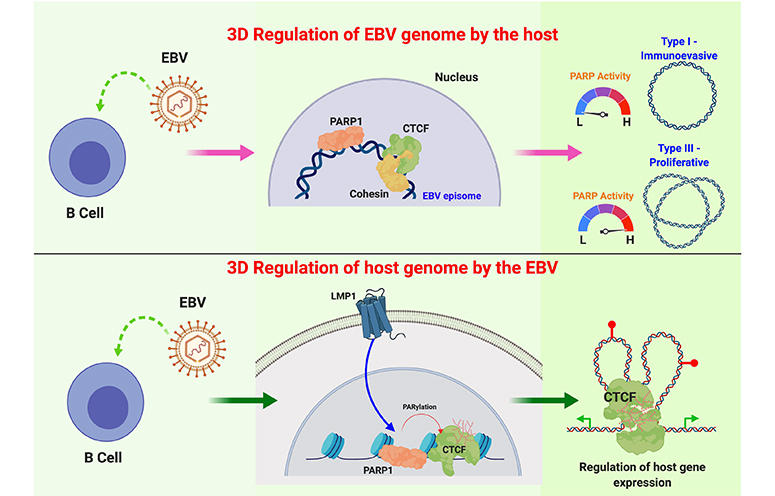
The Lieberman lab continues to study EBV and KSHV genome maintenance proteins, EBNA1 and LANA, respectively. These proteins bind to the viral OriP, but they also bind to the cellular chromosome at unknown sites. The Lieberman lab has identified the cellular chromosome binding sites for both EBNA1 and LANA in latently infected B-lymphocytes. LANA was found to bind to host genes involved in gamma-interferon signaling and LANA may antagonize STAT1/STAT3 binding to host genes important for MHC peptide presentation and processing. EBNA1 may promote higher order structures, including interchromosome linkages that may promote translocations similar to those observed in Burkitt’s lymphoma.
CHROMOSOME CONFORMATION CONTROL OF VIRAL LATENCY
The role of chromosome architecture and higher-ordered structure is also important for genome maintenance. The Lieberman lab has studied the role of chromatin architecture proteins CTCF and cohesins in regulating viral genome structure and gene expression during latent infection. They have shown that CTCF and cohesins mediate long-distance interactions that are important for control of gene expression and maintenance of a stable latent infection. Loss of genome architecture leads to a change in gene expression and a transition from a circular to linear viral genome.
TELOMERE MAINTENANCE AND DYSFUNCTION IN CANCER
Maintenance of telomere structures that maintain the ends of linear chromosomes is also important for human genome stability. The Lieberman lab is studying the chromatin structure of telomeres and the expression of a telomere repeat-containing non-coding RNA, termed TERRA. They have shown that TERRA is overexpressed in highly proliferating cells in human and mouse cancers. The TERRA form nuclear aggregates in cancer cells in mouse models of medulloblastoma, and TERRA RNA levels were highly over-expressed in human ovarian cancer biopsies. The regulation and function of TERRA expression, and its role in regulating telomere length and stability are the focus of future research.
DRUG DISCOVERY RESEARCH
The Lieberman laboratory is also pursuing the development of small molecule inhibitors of the EBV encoded origin binding protein EBNA1. The laboratory is collaborating with structural biologists and medicinal chemists to advance hits into lead compounds for testing in animal models of EBV lymphomagenesis. These small molecules will be considered for further development as inhibitors of EBV-associated malignancies.
Lieberman Lab in the News
Selected Publications
-
Cryo-EM Structure and Functional Studies of EBNA1 Binding to the Family of Repeats and Dyad Symmetry Elements of Epstein-Barr Virus oriP.
Mei, Y., Messick, T.E., Dheekollu, J., Kim, H.J., Molugu, S., Castro Muñoz, L.J.C., Moiskeenkova-Bell, V., Murakami, K., Lieberman, P.M. “Cryo-EM Structure and Functional Studies of EBNA1 Binding to the Family of Repeats and Dyad Symmetry Elements of Epstein-Barr Virus oriP.” J Virol. 2022 Sep 14;96(17):e0094922. doi: 10.1128/jvi.00949-22. Epub 2022 Aug 29.
-
DAXX-ATRX Regulation of p53 Chromatin Binding and DNA Damage Response.
Gulve, N., Su, C., Deng, Z., Soldan, S.S., Vladimirova, O., Wickramasinghe, J., Zheng, H., Kossenkov, A.V. , Lieberman, P.M. “DAXX–ATRX Regulation of p53 Chromatin Binding and DNA Damage Response.” Nat Commun. 2022 Aug 26;13(1):5033. doi: 10.1038/s41467-022-32680-8.
-
Epigenetic Plasticity Enables CNS-Trafficking of EBV-infected B Lymphocytes.
Soldan, S.S., Su, C., Lamontagne, R.J., Grams, N., Lu, F., Zhang, Y., Gesualdi, J.D., Frase, D.M., Tolvinski, L.E, Martinm, K., et al. “Epigenetic Plasticity Enables CNS-Trafficking of EBV-infected B Lymphocytes.” PLoS Pathog. 2021 Jun 9;17(6):e1009618. doi: 10.1371/journal.ppat.1009618. eCollection 2021 Jun.
-
A Multi-omics Approach to Epstein-Barr Virus Immortalization of B-cells Reveals EBNA1 Chromatin Pioneering Activities Targeting Nucleotide Metabolism.
Lamontagne, R.J., Samantha, S.S., Su, C., Wiedmer, A., Won, K.J., Lu, F., Goldman, A.R., Wickramasinghe, J., Tang, H.Y., Speicher, D.W., et al. “A Multi-omics Approach to Epstein-Barr Virus Immortalization of B-cells Reveals EBNA1 Chromatin Pioneering Activities Targeting Nucleotide Metabolism.” PLoS Pathog. 2021 Jan 26;17(1):e1009208. doi: 10.1371/journal.ppat.1009208. eCollection 2021 Jan.
-
Cell-cycle-dependent EBNA1-DNA Crosslinking Promotes Replication Termination at oriP and Viral Episome Maintenance.
Dheekollu, J., Wiedmer, A., Ayyanathan, K., Deakyne, J.S., Messick, T.E., Lieberman, P.M. “Cell-cycle-dependent EBNA1–DNA Crosslinking Promotes Replication Termination at oriP and Viral Episome Maintenance.” Cell. 2021 Feb 4;184(3):643-654.e13. doi: 10.1016/j.cell.2020.12.022. Epub 2021 Jan 21.